Welcome to lme4u
Hierarchical (or mixed-effects) models are essential tools for
analyzing data with grouped or nested structure — think students within
schools or patients within hospitals. And when it comes to fitting these
models in R, the lme4 package is the go-to hub.
While lme4 offers incredible modeling features, it can
also be hard to wrap your head around. Many users — especially those
transitioning from stats::lm() to lme4::lmer()
or even lme4::glmer() — find themselves overwhelmed by the
jump in complexity: random intercepts and slopes, link-scale estimates,
nested structures, and more. That’s where lme4u comes into
play.
Built directly on top of lme4, this package is designed to help you
interpret, diagnose, and
hypothesis-test your mixed models with clarity and
confidence. Whether you’re a student learning mixed models for the first
time or a researcher working with real-world grouped data,
lme4u walks you through the process with practical
explanations and inspiring tools.
Example Context: Students in Schools
The Data
lme4u comes with a built-in dataset, star,
a cleaned subset of the well-known Project
STAR (student-teacher achievement ratio) data. The original
dataset follows students across elementary school years in Tennessee,
aiming to study the effect of class size on student test performance.
The processed version in lme4u focuses on student
performance in third grade, ensuring a hierarchical structure where
students are nested within schools.
The shape of star is 4192 * 13, which includes a subset
of key variables related to student demographics, prior-year (2nd grade)
and current-year (3rd grade) test scores, class assignment, teacher
qualifications, and school-level identifiers. All students in this
dataset have been controlled as attending the same school in both 2nd
and 3rd grades. To view details and data dictionary, run
?star after loading lme4u into your
workspace.
The Model
Under the context of star, suppose we’re interested in
understanding:
How does class type affect student math performance, after accounting for prior achievements? And does this relationship vary across schools?
This type of question is common in education research, where students
are grouped within schools, and both individual- and group-level
heterogeneity contribute to outcomes. In R using lme4, this
would be implemented as:
model <- lmer(math ~ cltype + math_old + (cltype | school_id), data = star)Of course, there could be other ways of modeling the relation. But we
will use this specific model to demonstrate how lme4u makes
interpretation and diagnostics easier throughout the vignette.
Interpreting the Model
Immediately after the model fit, you might start wondering whether
the model structure is appropriate, or simply, what the formula means
statistically. A core component of the package,
explain_lmer(), provides structured, user-friendly
interpretations of linear mixed effects models fitted using
lme4::lmer(). This function is designed to bridge the gap
between raw model output and intuitive understanding, making it easier
for users to interpret their model structure and results.
General Summary
explain_lmer() supports three levels of interpretation
via the details argument. By default,
"general" will be run if no argument supplied to
details, which provides a high-level summary of the model,
describing its structure, fixed effects, and random effects in a concise
yet informative manner:
explain_lmer(model)
#> This linear mixed-effects model examines how cltype, math_old affects math, while accounting for group-level variability across school_id.
#>
#> The model adjusts for fixed effect(s), including cltype, math_old, which estimate the overall relationship with math across the entire dataset. For more detailed interpretations of fixed effects, use `details = "fixed"`.
#>
#> The model includes the following random effects across grouping variables: cltype | school_id.
#> *The random intercept(s), school_id, account for variations in the intercept across different groups, allowing each group to have its own baseline value that deviates from the overall average.
#> *On top of the varying baselines, the model allows the effects of cltype on math to differ across their corresponding groups. This means that not only can each group start from a different baseline, but the strength and direction of the relationships between these variables and math can also vary from one group to another.
#> Check `details = 'random'` for more interpretation.Fixed Effects
If "fixed" is input, the function offers a detailed
explanation of the fixed effects, including interpretations of
individual estimates and their implications within the model
context:
fix <- explain_lmer(model, details = "fixed")
#> This model includes the following fixed effects: cltypesmall, cltyperegular+aide, math_old, along with the baseline estimate (Intercept).
#> Fixed effects estimate the average relationship between the predictors and the response variable `math` across all groups.
#> These effects reflect how changes in each predictor influence `math`, assuming all other variables are held constant and deviations from the baseline are measured accordingly.
#>
#> To interpret the estimates:
#> - The <Estimate> represents the expected change in `math` for a one-unit change in the predictor.
#> - The <Standard Error> reflects uncertainty around the estimate.
#> - The <t-value> measures the significance of the effect relative to its variability.
#> - The <Correlation> provides insight into the relationships between fixed effect predictors.
#>
#>
#> For detailed interpretations of each fixed effect, use: `attr(, 'fixed_details')[['variable_name']]` to view specific results. You can find the list of available fixed effect variable names using: `attr(, 'fixed_names')` to guide your extraction.
attr(fix, "fixed_details")
#> $`(Intercept)`
#> [1] "The intercept estimate suggests that when all predictors are set to zero, the expected value of `math` is approximately 201.646. This serves as the baseline for evaluating the effects of other variables in the model."
#>
#> $cltypesmall
#> [1] "The fixed effect for 'cltypesmall' suggests that for a one-unit increase in 'cltypesmall', the expected change in `math` is approximately -0.434, assuming all other variables are held constant. This estimate has a standard error of 2.406 and a t-value of -0.18. To assess the overall impact of this effect, consider it relative to the model's baseline intercept."
#>
#> $`cltyperegular+aide`
#> [1] "The fixed effect for 'cltyperegular+aide' suggests that for a one-unit increase in 'cltyperegular+aide', the expected change in `math` is approximately -0.117, assuming all other variables are held constant. This estimate has a standard error of 2.503 and a t-value of -0.047. To assess the overall impact of this effect, consider it relative to the model's baseline intercept."
#>
#> $math_old
#> [1] "The fixed effect for 'math_old' suggests that for a one-unit increase in 'math_old', the expected change in `math` is approximately 0.715, assuming all other variables are held constant. This estimate has a standard error of 0.01 and a t-value of 69.735. To assess the overall impact of this effect, consider it relative to the model's baseline intercept."NOTE: If interaction is fitted to the model, the function is also able to detect and adjust the interpretation.
Random Effects
If "random" is input, the function focuses on random
effects, explaining random intercepts, slopes, and variance components
to highlight across-group variability:
random <- explain_lmer(model, details = "random")
#> There is 1 random term you fit in the model:
#>
#> cltype | school_id: Your model assumes there is across-group heterogeneity in the baseline level of math across school_id (random intercepts), allowing each group to have its own starting point that deviates from the overall average. Additionally, the effects of cltype on math are allowed to vary within each school_id (random slopes), capturing differences in these relationships across groups.
#>
#> To explore variance contributions based on the model estimates, use: `attr(, 'var_details')`.
attr(random, "var_details")
#> $high_var
#> [1] "The following terms explain a substantial proportion of the variance: Residual. This suggests that these random effects contribute meaningfully to capturing group-level heterogeneity. Note: The residual variance dominates, suggesting that within-group heterogeneity explains most of the variation. This could indicate a weak random effect structure."
#>
#> $low_var
#> [1] "The following terms explain a relatively small portion of the variance: NA, school_id - (Intercept). You might reconsider including these terms in your random effect structure if their contributions are negligible."NOTE: The random part is also capable of detecting nested grouping structure, uncorrelated random effects, and multi-level random structures. However, use the interpretation deliberately under such complex cases.
Assumption Diagnostics
Understanding the model structure is just a step-one. Now the critical question shifts to whether the model assumptions are reasonably satisfied. In the context of linear mixed-effect models, assumption diagnostics usually consider the following:
- Within-Group Variation
- Residuals are independent
- Residuals have equal variance in each group (homoscedasticity)
- Residuals follow a roughly normal distribution
- Between-Group Variation
- Group-specific effects (e.g., intercepts or slopes) are independent
- Group-specific effects follow a roughly normal distribution
Using the suite of residuals functions in lme4u
(res_norm(), res_fit(),
res_box()), you could gain insights into the within-group
heterogeneity and detect possible violations through the plots. Note
that due to the varying nature of complex random structures,
between-group assumptions are not included in the package.
Normality of Residuals
From the QQ plot, we can see the residual dots generally follow the normality line, suggesting a meet of the assumption:
res_norm(model)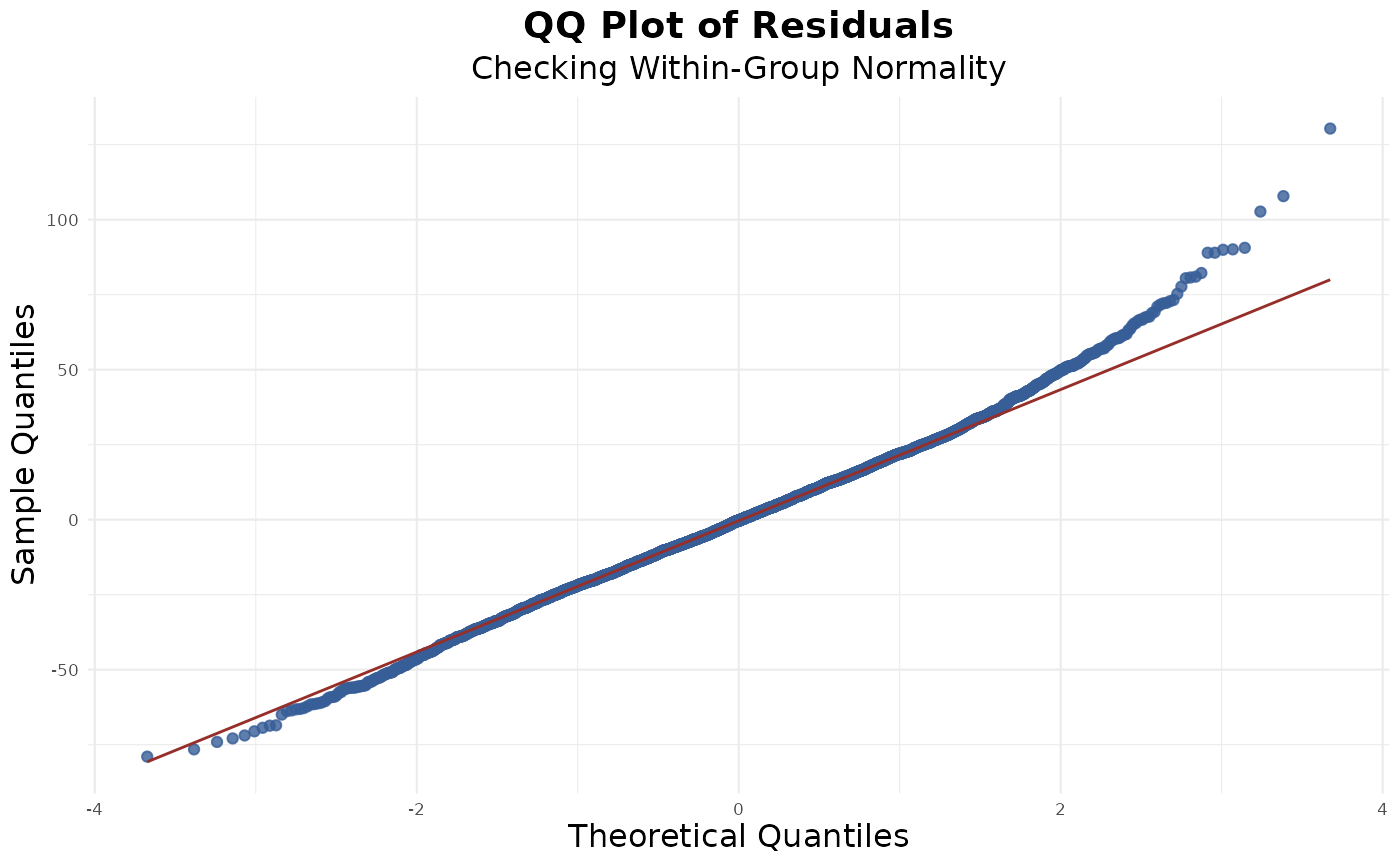
Mean-Variance Relationship
The residuals vs. fitted plot does not exhibit the classic funnel-shaped pattern, suggesting that the assumption of constant variance (homoscedasticity) is reasonably met. Consider variable transformations if you do notice funneling or specific patterns:
res_fit(model)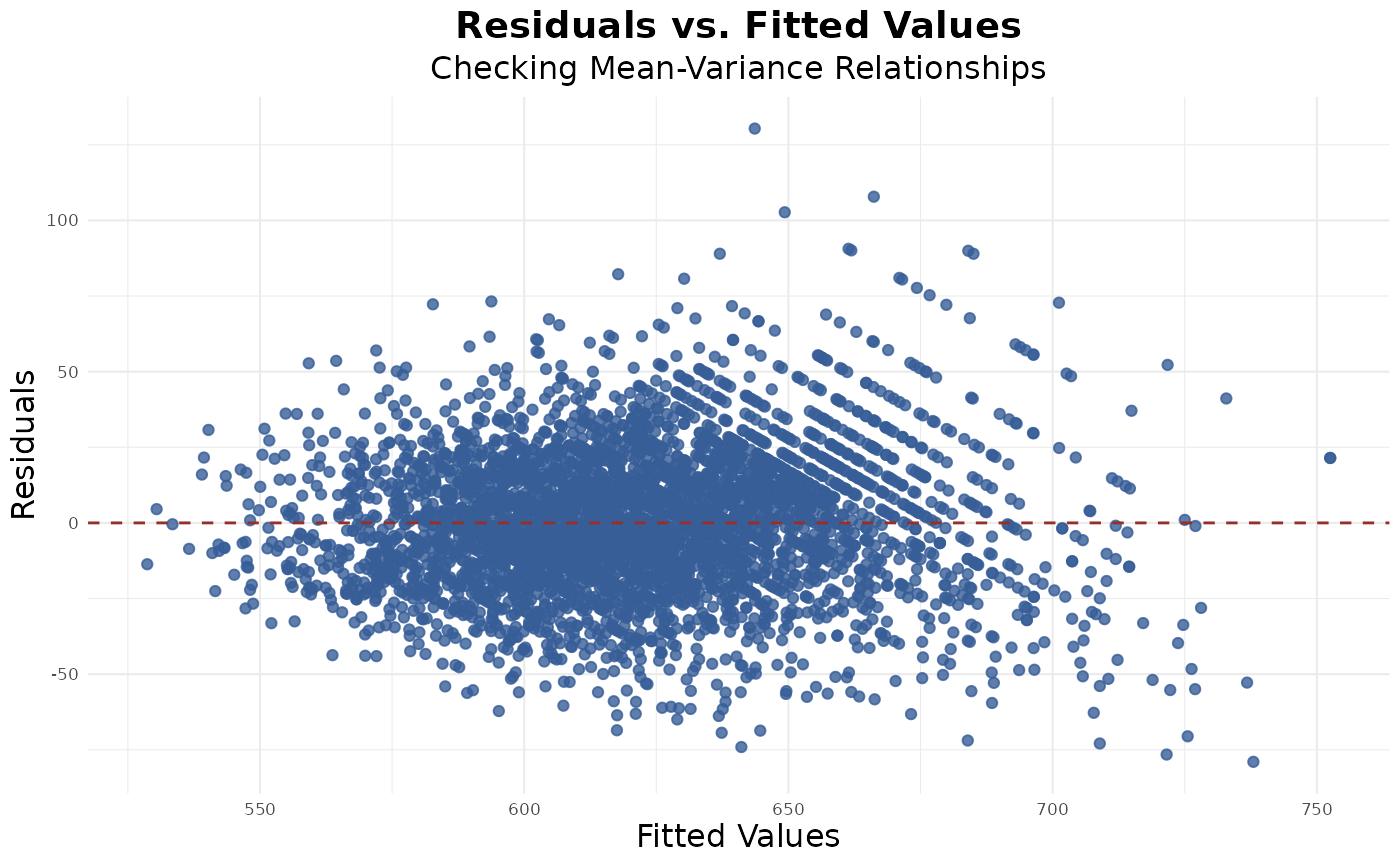
Homoscedasticity
The boxplot of residuals by school shows relatively consistent spread across most groups, suggesting that the assumption of equal residual variance within groups is reasonably satisfied:
res_box(model, group_var = "school_id")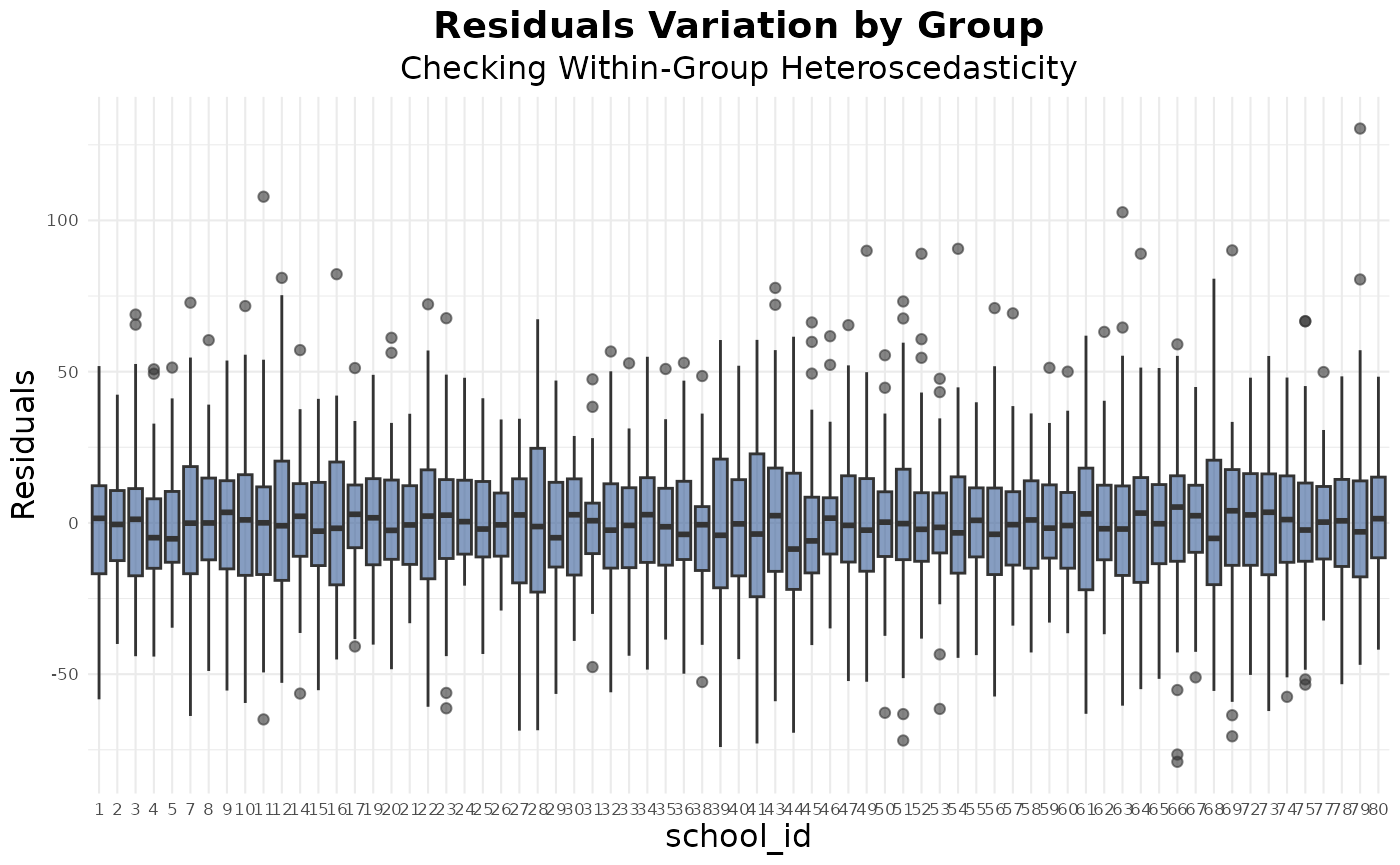
Testing Effects
Once you’ve interpreted your model and checked that assumptions
reasonably hold, the next natural step is to test whether certain
effects — fixed or random — meaningfully contribute to your model.
lrt_lmer() provides likelihood ratio tests on a selected
fixed or random effect in a fitted lme4::lmer() model. It
aims to gauge the effect brought in by one single term and apply
statistically accurate null distribution based on different effect
types.
Depending on whether you’re testing a fixed effect or a random effect
by the type argument, the structure of the reduced model
differs. Currently, the function only supports lmer models with fixed
effects and a single random structure (i.e., no nested
/, double vertical ||, or multi-level random
effects). To see more about the testing logic and how the reduced models
are constructed differently, run ?lrt_lmer.
Fixed Effects
To test whether a selected fixed effect exists, specify the exact
variable name in target and set
type = "fixed". This will allow the function to match with
the underlying set of fixed terms. Running the function will give a
summary of output, along with the test statistics being returned as a
list:
fix_result <- lrt_lmer(model, target = "math_old", type = "fixed", data = star)
#> Likelihood Ratio Test for 'math_old' (fixed effect)
#> Full Model: math ~ cltype + math_old + (cltype | school_id)
#> Reduced Model: math ~ cltype + (cltype | school_id)
#> Test Statistics:
#> - Log-Likelihood Difference: 3202.807
#> - Degrees of Freedom: 1 (Difference in estimated parameters)
#> - P-Value: < 2.2e-16 (Chi-square test)
#> Since p = < 2.2e-16, under alpha level of 0.05, we reject the null hypothesis. The test suggests that the fixed effect of math_old is statistically significant and contributes to the model.
fix_result
#> $logLik
#> [1] 3202.807
#>
#> $df
#> [1] 1
#>
#> $pvalue
#> [1] 0NOTE: If interaction is fitted to the model, specify
has_interaction = TRUE in the input argument. The reduced
model will be subsequently constructed without the selected fixed effect
and any interaction terms containing it.
Random Effects
To test the excess heterogeneity contributed by a random effect term,
specify the exact variable name in target and set
type = "random". Then, the function will check whether the
target is a random intercept (grouping variable) or a random slope in
the model, and apply corresponding computation for the likelihood ratio
test.
Random Intercept: The full model is
reconstructed by modifying the original lme4::lmer() call
to retain only the random intercept (ignoring any random slopes). The
reduced model is a standard stats::lm() model containing
only the fixed effects, and the likelihood ratio test follows a mixture
Chi-square distribution for p-value computation.
random_result1 <- lrt_lmer(model, target = "school_id", type = "random", data = star)
#> Likelihood Ratio Test for 'school_id' (random intercept effect)
#> Full Model: math ~ cltype + math_old + (1 | school_id)
#> Reduced Model: math ~ cltype + math_old
#> Test Statistics:
#> - Log-Likelihood Difference: 603.7954
#> - Degrees of Freedom: 1 (Difference in estimated parameters)
#> - P-Value: < 2.2e-16 (Chi-square test)
#> Since p = < 2.2e-16, under alpha level of 0.05, we reject the null hypothesis. The test suggests that the random intercept effect of school_id is statistically significant and contributes to the model.Random Slope: The target variable is only
removed from the random slope structure. The reduced model remains an
lme4::lmer() model, and the likelihood ratio test follows a
mixture Chi-square distribution for p-value computation.
random_result2 <- lrt_lmer(model, target = "cltype", type = "random", data = star)
#> Likelihood Ratio Test for 'cltype' (random slope effect)
#> Full Model: math ~ cltype + math_old + (cltype | school_id)
#> Reduced Model: math ~ cltype + math_old + (1 | school_id)
#> Test Statistics:
#> - Log-Likelihood Difference: 326.1249
#> - Degrees of Freedom: 5 (Difference in estimated parameters)
#> - P-Value: < 2.2e-16 (Chi-square test)
#> Since p = < 2.2e-16, under alpha level of 0.05, we reject the null hypothesis. The test suggests that the random slope effect of cltype is statistically significant and contributes to the model.